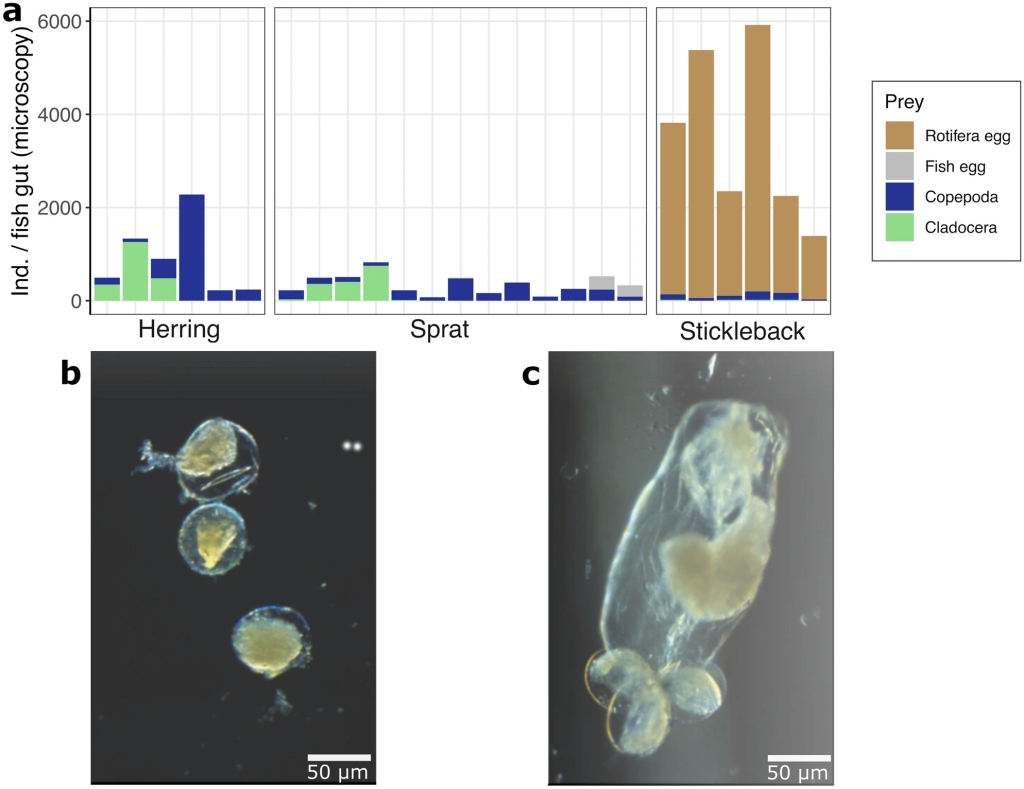
Here, we investigate diet overlap between these three planktivorous fishes in the Baltic Sea, utilizing DNA metabarcoding on the 18S rRNA gene and the COI gene, targeted qPCR, and microscopy. Our results show niche differentiation between clupeids and stickleback, and highlight that rotifers play an important role in this pattern, as a resource that is not being used by the clupeids nor by other zooplankton in spring. We further show that all the diet assessment methods used in this study are consistent, but also that DNA metabarcoding describes the plankton-fish link at the highest taxonomic resolution. This study suggests that rotifers and other understudied soft-bodied prey may have an important function in the pelagic food web and that the growing population of pelagic stickleback may be supported by the open feeding niche offered by the rotifers.
The study is available here: Novotny A, Jan KMG, et al. Scientific Reports 2022