A new study shows that resource use overlaps among herring, sprat and three-spined stickleback are limited in the Baltic Sea and varied with prey availability. We assessed diet composition in May and October using DNA metabarcoding, stable isotopes, and microscopy. All three methods gave consistent results.
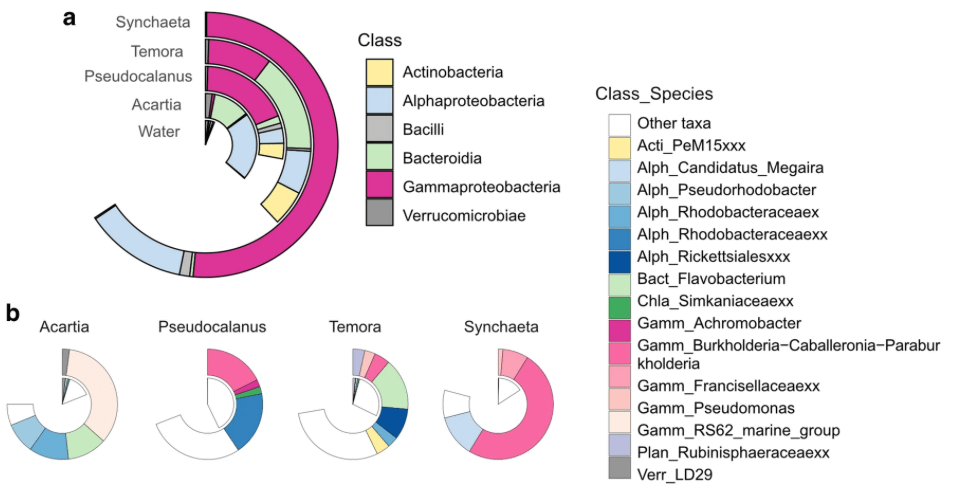
The clupeids shared a similar diet in May when prey diversity was low, composed mainly of the copepods Pseudocalanus and Acartia, whereas three-spined stickleback favoured different copepod species and the rotifer Synchaeta, which was confirmed by a different isotopic value as compared to the two clupeids. In October, all forage fish preyed on diverse zooplankton species, mainly composed of the copepods Acartia, Eurytemora, and Temora, while Pseudocalanus was only important for herring. The observed resource use partitioning between sprat and herring was confirmed by the stable isotope values from October, suggesting that different prey species were targeted during the summer period.
This study suggests that shifts in zooplankton dynamics, rather than competition for resources, have the potential to drive small pelagic fish population fluctuations.
Find the full publication at:
Kinlan M G Jan, Jonas Hentati-Sundberg, Niklas Larson, Monika Winder, Limited resource use overlaps among small pelagic fish species in the central Baltic Sea, ICES Journal of Marine Science, Volume 82, Issue 9, September 2025, fsaf122, https://doi.org/10.1093/icesjms/fsaf122

Figure 4. Prey selectivity for herring, sprat, and three-spined stickleback. The link widths are proportional to the average Chesson index for each fish species in May (a, b) and October (c, d) based on the 18S (a, c) and COI (b, d) reads. Asterisks represent prey taxa observed only in one season for each barcode.